4 Infrared Thermography
The development of a non-invasive tool (infrared thermography) to measure stress and pain in animals has significant applications in assuring the welfare standards of the New Zealand agricultural industry and boosting the value of its products in overseas markets. Here we are interested in stress involved in the disbudding (removal of horns) of cattle.
This lesson investigates the emotional and or stress responses of cattle during disbudding using four different treatment options. The original research was carried out by Mairi Stewart (AgResearch, Ruakura).
Data
There is 1 file associated with this presentation. It contains the data you will need to complete the lesson tasks.
Video
Objectives
Tasks
0. Read and Format data
0a. Read in the data
First check you have installed the package readxl (see Section 2.6) and set the working directory (see Section 2.1), using instructions in Getting started with R.
Load the data into R.
This creates a data frame containing data from a study investigating the effects of 4 different disbudding treatments on dairy calves.
The code has been hidden initially, so you can try to load the data yourself first before checking the solutions.
Code
#loads readxl package
library(readxl)
#loads the data file and names it disbud
disbud<-read_xls("Disbud stress.xls")
#view beginning of data frame
head(disbud)Code
#loads readxl package
library(readxl) Warning: package 'readxl' was built under R version 4.2.2Code
#loads the data file and names it disbud
disbud<-read_xls("Disbud stress.xls")New names:
• `` -> `...7`
• `` -> `...8`
• `` -> `...9`
• `` -> `...10`
• `` -> `...11`
• `` -> `...12`
• `` -> `...13`Code
#view beginning of data frame
head(disbud)# A tibble: 6 × 13
Tag `Trt num` Trt Base e…¹ Tempe…² Chang…³ ...7 ...8 ...9 ...10 ...11
<dbl> <dbl> <chr> <dbl> <dbl> <dbl> <lgl> <chr> <chr> <chr> <chr>
1 5124 1 Control 37.0 37.2 0.212 NA Trea… <NA> <NA> <NA>
2 5659 1 Control 37.1 36.8 -0.303 NA DB Disb… <NA> <NA>
3 5656 1 Control 37.4 37.6 0.238 NA LADB Disb… <NA> <NA>
4 5677 1 Control 37.9 37.8 -0.0871 NA Cont… Sali… <NA> <NA>
5 5132 1 Control 37.6 37.8 0.261 NA LA Loca… <NA> <NA>
6 5950 1 Control 38.1 37.9 -0.157 NA <NA> <NA> <NA> <NA>
# … with 2 more variables: ...12 <chr>, ...13 <chr>, and abbreviated variable
# names ¹`Base eye temperature (°C)`,
# ²`Temperature in first 3 mins post treatment (°C)`,
# ³`Change in eye temperature (°C)`
# ℹ Use `colnames()` to see all variable namesIn this study 30 calves were allocated to one of 4 treatments for disbudding then stress levels were monitored using infrared thermography (a non-invasive method which measured eye-temperature).
The variables recorded are Tag, Trt num (treatment number), Trt(treatment),Base eye temperature (°C), Temperature in first 3 mins post treatment (°C), and Change in eye temperature (°C). The four treatment types recorded are Control, LA (local anaesthetic injection, mock disbudding), DB (disbudded with saline injection) and LADB (disbudded with local anaesthetic injection).
0b. Format the data
Tidy up the data frame by specifying Trt as a factor variable and renaming some of the variables for easier reference later.
Code
#reassign as factor variable
disbud$Trt<-as.factor(disbud$Trt)
#rename columns for easier reference
colnames(disbud)<-c("Tag","TrtNum","Trt","BaseEyeTemp","PostTreatEyeTemp","ChangeEyeTemp") Code
#reassign as factor variable
disbud$Trt<-as.factor(disbud$Trt)
#rename columns for easier reference
colnames(disbud)<-c("Tag","TrtNum","Trt","BaseEyeTemp","PostTreatEyeTemp","ChangeEyeTemp") Another data tidying step we can take is removing extra columns and NAs.
Code
#overwrite the original disbud data frame with the data frame without NAs using na.omit()
#subsetting with [] to only include the first 6 columns, the remaining 7 have comments/information we do not need
disbud<-na.omit(disbud[,c(1:6)]) Code
#overwrite the original disbud data frame with the data frame without NAs using na.omit()
#subsetting with [] to only include the first 6 columns, the remaining 7 have comments/information we do not need
disbud<-na.omit(disbud[,c(1:6)]) 1. Experimental Design
Discuss the importance of a randomised design.
How many treatments are present?
Why might one of the treatments have only 6 cattle?
Is there a control treatment that other treatments are compared with?
See video Section 4.1
A randomised design is important as it controls the effects of potential confounding variables (in the present study this could include factors such as breed of calf, temperament). This allows researchers to detect a true effect/relationship between the variables of interest if one does exist.
There were 4 treatments present in this study - handling only, local anaesthetic only, disbudding only, and disbudding with local anaesthetic.
The disbudding only treatment group had only 6 cattle as this was the minimum number required to detect a true difference between groups (with 30 calves across groups). As disbudding without local anaesthetic was the most painful treatment group, for ethical reasons the number of calves were limited here.
There were 2 control treatments that the others were compared with. These were handling only and local anaesthetic only.
2. Box Plot, Scatter Plot
It is important to know what our data looks like, as it may help us to understand the results of the calculations that we will be doing later on.
Plot ChangeEyeTemp against the four Trt groups.
What can you see from the graphs?
Code
#plot() function returns a box plot for each treatment as this is a categorical variable
plot(disbud$Trt, disbud$ChangeEyeTemp) Have a look at the help file for the plot() function to figure out how to produce a scatter plot instead.
Code
#plot() function returns a box plot for each treatment as this is a categorical variable
plot(disbud$Trt, disbud$ChangeEyeTemp)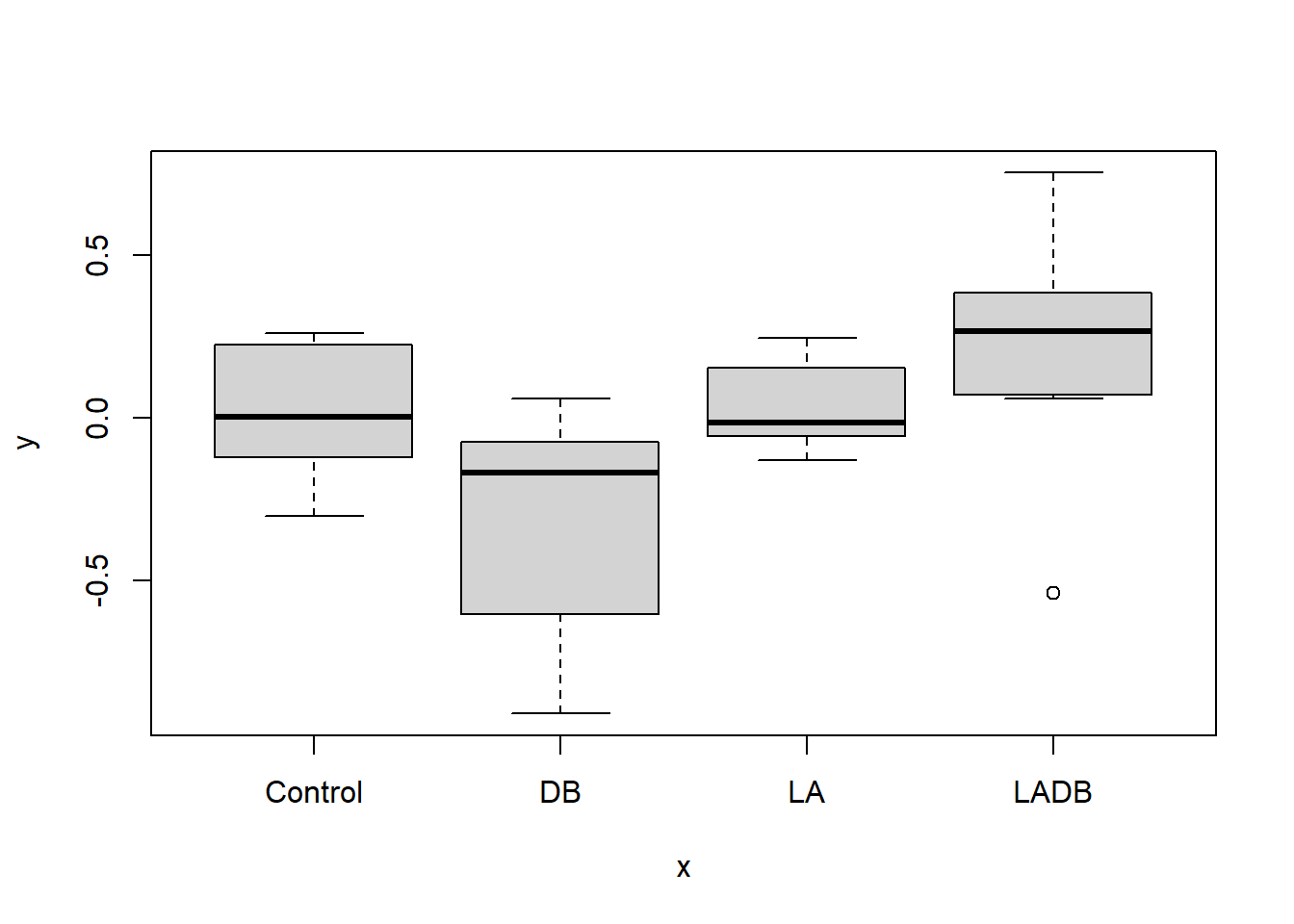
Code
#can see individual points using plot.default() instead
plot.default(disbud$Trt, disbud$ChangeEyeTemp)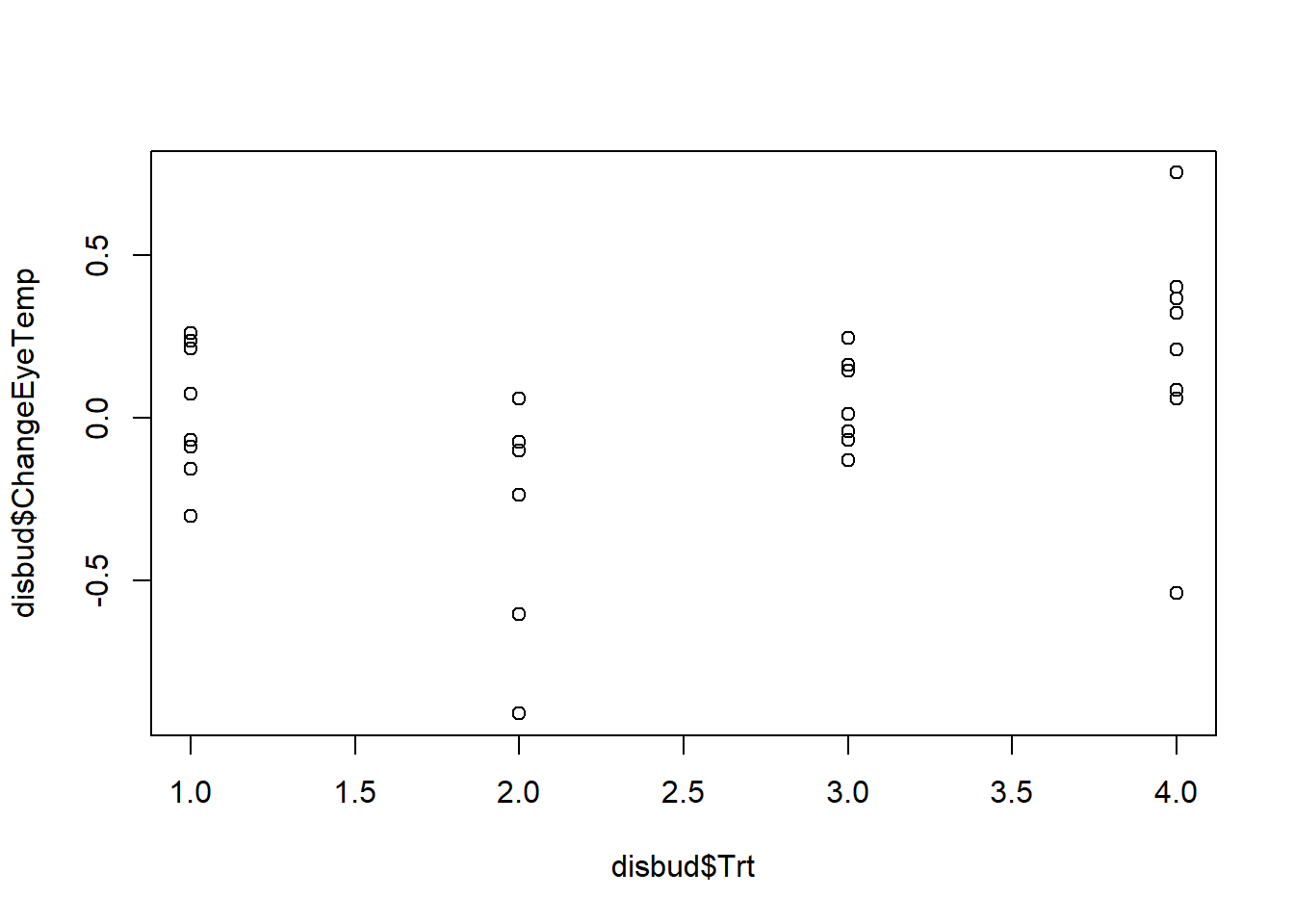
3. Confidence Interval (mean)
Calculate the mean ChangeEyeTemp and the standard error for each Trt mean (SEM).
Using these summary statistics and the formula below, calculate a 95% confidence interval for the mean change in eye temperature for each treatment group (use a large sample value of 1.96 or 2).
\[ \overline{x} \pm 1.96 * \frac{\sigma_x}{\sqrt{n}}\]
What do you conclude?
Code
#estimate of the mean change in eye temperature for control cattle
mean(disbud$ChangeEyeTemp[disbud$Trt=="Control"])
#estimate of the standard error of the mean change in eye temperature for control cattle
sd(disbud$ChangeEyeTemp[disbud$Trt=="Control"])/sqrt(length(disbud$ChangeEyeTemp[disbud$Trt=="Control"])) Calculate these components for the LA, DB, and LADB Trt groups.
Code
#eye temperature for control cattle
m<-mean(disbud$ChangeEyeTemp[disbud$Trt=="Control"])
se<-sd(disbud$ChangeEyeTemp[disbud$Trt=="Control"])/sqrt(length(disbud$ChangeEyeTemp[disbud$Trt=="Control"]))
m-1.96*se[1] -0.1220748Code
m+1.96*se[1] 0.1647258Code
#eye temperature for local anaesthetic cattle
m<-mean(disbud$ChangeEyeTemp[disbud$Trt=="LA"])
se<-sd(disbud$ChangeEyeTemp[disbud$Trt=="LA"])/sqrt(length(disbud$ChangeEyeTemp[disbud$Trt=="LA"]))
m-1.96*se[1] -0.05590293Code
m+1.96*se[1] 0.1273838Code
#eye temperature for disbudding cattle
m<-mean(disbud$ChangeEyeTemp[disbud$Trt=="DB"])
se<-sd(disbud$ChangeEyeTemp[disbud$Trt=="DB"])/sqrt(length(disbud$ChangeEyeTemp[disbud$Trt=="DB"]))
m-1.96*se[1] -0.6067618Code
m+1.96*se[1] -0.0143709Code
#eye temperature for disbudding and local anaesthetic cattle
m<-mean(disbud$ChangeEyeTemp[disbud$Trt=="LADB"])
se<-sd(disbud$ChangeEyeTemp[disbud$Trt=="LADB"])/sqrt(length(disbud$ChangeEyeTemp[disbud$Trt=="LADB"]))
m-1.96*se[1] -0.04967224Code
m+1.96*se[1] 0.4657519In practice a t-value (calculated using the function qt()) should be used for confidence intervals as we have small samples for each of the treatments. The t-value would have 8-1=7 degrees of freedom (df) for treatments 1, 2, and 4, but only 6-1 df for treatment 3.
The function t.test() carries out this entire process automatically. You can try this for an extension.
4. Bootstrap Confidence Interval, Histogram (mean)
For each Trt, we want to calculate the bootstrap confidence interval for the mean ChangeEyeTemp.
The bootstrap technique involves randomly sampling a new dataset from the original sampled data. This creates a large number of datasets, that may have been seen if the experiment had been repeated many times (i.e. creates new datasets based on the original dataset).
This can be done in R using the boot package.
What distribution do the means of the bootstrap samples appear to follow?
How do the bootstrap estimates of the mean for each treatment group compare to the treatment means we calculated previously? Do the bootstrap confidence intervals lead to the same conclusions as before (for each treatment group)?
Code
library(boot)
#set seed so you get the same results when running again
set.seed(4)
#function that calculates the mean from the bootstrapped samples
meanFun<-function(data,indices){
dataInd<-data[indices,] #sample rows with replacement from the data
meandata<-mean(dataInd$ChangeEyeTemp) #calculate statistic of interest, in this case the mean, from the sample
return(meandata)
}
#use the boot() function to bootstrap mean change in eye temp for control cattle
#create object called meanBoot with the output of the bootstrapping procedure
meanBoot<-boot(disbud[disbud$Trt=="Control",],statistic=meanFun, R=10000) After running the bootstrap sampling, we can view summary statistics of the results by calling the object.
Code
meanBootFrom the \(\verb!Bootstrap Statistics!\) output we can calculate a normal 95% confidence interval using the estimate of \(\verb!t1*!\) and the \(\verb!std. error!\).
Code
lower<-0.02132551-1.96*0.06804017
upper<-0.02132551+1.96*0.06804017It is also useful to produce a graph of the distribution of the results.
This graph shows the distribution of the mean of the bootstrap samples with a density curve and 95% confidence interval limits plotted on top.
Code
#plot density (freq=F) histogram of bootstrap results
hist(meanBoot$t,freq=F)
#add a smoothed line to show the distribution
lines(density(meanBoot$t))
#add vertical lines with lower and upper confidence limits
abline(v=c(lower,upper))Code
library(boot)Warning: package 'boot' was built under R version 4.2.2Code
#set seed so you get the same results when running again
set.seed(4)
#function that calculates the mean from the bootstrapped samples
meanFun<-function(data,indices){
dataInd<-data[indices,] #sample rows with replacement from the data
meandata<-mean(dataInd$ChangeEyeTemp) #calculate statistic of interest, in this case the mean, from the sample
return(meandata)
}
#use the boot() function to bootstrap mean change in eye temp for control cattle
#create object called meanBoot with the output of the bootstrapping procedure
meanBoot<-boot(disbud[disbud$Trt=="Control",],statistic=meanFun, R=10000) After running the bootstrap sampling, we can view summary statistics of the results by calling the object.
Code
meanBoot
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot(data = disbud[disbud$Trt == "Control", ], statistic = meanFun,
R = 10000)
Bootstrap Statistics :
original bias std. error
t1* 0.02132551 -8.071731e-05 0.06858631From the \(\verb!Bootstrap Statistics!\) output we can calculate a normal 95% confidence interval using the estimate of \(\verb!t1*!\) and the \(\verb!std. error!\).
Code
lower<-0.02132551-1.96*0.06804017
upper<-0.02132551+1.96*0.06804017
lower[1] -0.1120332Code
upper[1] 0.1546842It is also useful to produce a graph of the distribution of the results.
This graph shows the distribution of the mean of the bootstrap samples with a density curve and 95% confidence interval limits plotted on top.
Code
#plot density (freq=F) histogram of bootstrap results
hist(meanBoot$t,freq=F)
#add a smoothed line to show the distribution
lines(density(meanBoot$t))
#add vertical lines with lower and upper confidence limits
abline(v=c(lower,upper))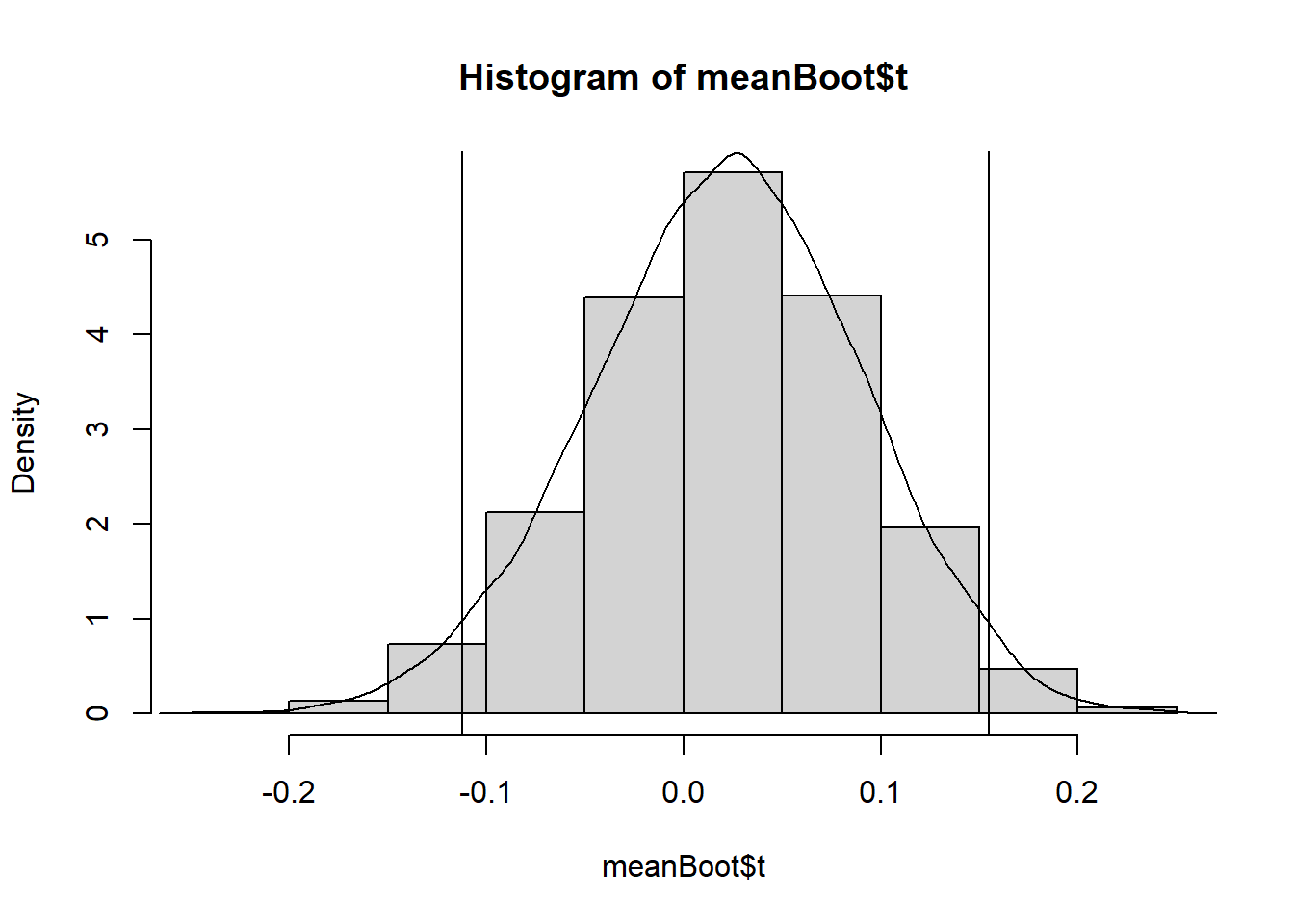
The means of bootstrapped samples follow a normal distribution, symmetric around a mean of 0.02132551. This mean of means matches the mean of the control treatment group we calculated earlier, and the distribution of bootstrapped mean estimates around it give an indication of the amount of variability in our estimate due to sampling.
The bootstrap confidence interval is similar to the one calculated earlier and includes both positive and negative values so leads to the same conclusion. There is no evidence to conclude a significant change in mean eye temperature for control cattle.
Repeat the bootstrapping above for the other Trt groups, and produce histograms of their results.
How do the bootstrap estimates of the mean for each treatment group compare to the treatment means we calculated previously? Do the bootstrap confidence intervals lead to the same conclusions as before (for each treatment group)?
5. Confidence Interval (difference in means)
To compare the Trt means for the DB and LABD treatments we want to set up a 95% confidence interval for the difference between the means using an unpaired t.test().
What conclusion can be drawn from our confidence intervals in regard to the effect of the local anaesthetic?
Code
#first test if variances are equal
var.test(disbud$ChangeEyeTemp[disbud$Trt=="DB"], disbud$ChangeEyeTemp[disbud$Trt=="LADB"],
alternative = "two.sided")
#no significant evidence against null hypothesis that variances are equal, use var.equal=T in t test
#compare the change in eye temperature for disbudding with saline and disbudding with local anaesthetic.
#paired=FALSE indicates independent samples.
t.test(disbud$ChangeEyeTemp[disbud$Trt=="DB"],disbud$ChangeEyeTemp[disbud$Trt=="LADB"],
paired=FALSE,var.equal=T) Calculate the difference in means using the \(\verb!sample estimates:!\) section of the output above.
Code
#first test if variances are equal
var.test(disbud$ChangeEyeTemp[disbud$Trt=="DB"], disbud$ChangeEyeTemp[disbud$Trt=="LADB"],
alternative = "two.sided")
F test to compare two variances
data: disbud$ChangeEyeTemp[disbud$Trt == "DB"] and disbud$ChangeEyeTemp[disbud$Trt == "LADB"]
F = 0.99071, num df = 5, denom df = 7, p-value = 0.9712
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.1874494 6.7894420
sample estimates:
ratio of variances
0.9907146 Code
#no significant evidence against null hypothesis that variances are equal, use var.equal=T in t test
#compare the change in eye temperature for disbudding with saline and disbudding with local anaesthetic.
#paired=FALSE indicates independent samples.
t.test(disbud$ChangeEyeTemp[disbud$Trt=="DB"],disbud$ChangeEyeTemp[disbud$Trt=="LADB"],
paired=FALSE,var.equal=T)
Two Sample t-test
data: disbud$ChangeEyeTemp[disbud$Trt == "DB"] and disbud$ChangeEyeTemp[disbud$Trt == "LADB"]
t = -2.5871, df = 12, p-value = 0.02378
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.95536854 -0.08184378
sample estimates:
mean of x mean of y
-0.3105663 0.2080398 Code
#difference in means
-0.3105663-0.2080398[1] -0.5186061Construct confidence intervals for the other four differences in means using t.test().
6. Bootstrap Confidence Interval, Histogram (difference in means)
Calculate the bootstrap confidence interval for the difference between the two Trt means in Task 5.
How does the bootstrap estimate of the mean difference between DB and LADB compare with the estimate of the mean difference found in Task 5? Does the bootstrap confidence interval lead to the same conclusion as before?
Was this outcome expected?
Code
library(boot)
#set seed so you get the same results when running again
set.seed(16)
#function that calculates the difference in means from the bootstrapped samples
meandiffFun<-function(data,indices){
data1<-data[data$Trt=="DB",] #subset data by treatment levels, then sample from these
dataInd1<-data1[indices,]
data2<-data[data$Trt=="LADB",] #subset data by treatment levels, then sample from these
dataInd2<-data2[indices,]
meandiff<-mean(dataInd1$ChangeEyeTemp,na.rm=T)-mean(dataInd2$ChangeEyeTemp,na.rm=T) #calculate difference in means for the samples. Use na.rm=T to remove NAs
return(meandiff)
}
#bootstrap difference in mean change in eye temp for DB and LADB cattle.
#create object called meandiffBoot with the output of the bootstrapping procedure
meandiffBoot<-boot(disbud[disbud$Trt=="DB"|disbud$Trt=="LADB",],statistic=meandiffFun, R=10000) Code
#plot density histogram of bootstrap results
hist(meandiffBoot$t,freq=F)
#add a smoothed line to show the distribution
lines(density(meandiffBoot$t,na.rm=T)) View the summary output and calculate the confidence interval, then add this to your plot above using the abline() function).
Notice that the bootstrap mean and confidence interval are both negative. What causes the negative difference between the means and could we make it a positive difference?
Code
library(boot)
#set seed so you get the same results when running again
set.seed(16)
#function that calculates the difference in means from the bootstrapped samples
meandiffFun<-function(data,indices){
data1<-data[data$Trt=="DB",] #subset data by treatment levels, then sample from these
dataInd1<-data1[indices,]
data2<-data[data$Trt=="LADB",] #subset data by treatment levels, then sample from these
dataInd2<-data2[indices,]
meandiff<-mean(dataInd1$ChangeEyeTemp,na.rm=T)-mean(dataInd2$ChangeEyeTemp,na.rm=T) #calculate difference in means for the samples. Use na.rm=T to remove NAs
return(meandiff)
}
#bootstrap difference in mean change in eye temp for DB and LADB cattle.
#create object called meandiffBoot with the output of the bootstrapping procedure
meandiffBoot<-boot(disbud[disbud$Trt=="DB"|disbud$Trt=="LADB",],statistic=meandiffFun, R=10000) Code
#plot density histogram of bootstrap results
hist(meandiffBoot$t,freq=F)
#add a smoothed line to show the distribution
lines(density(meandiffBoot$t,na.rm=T)) 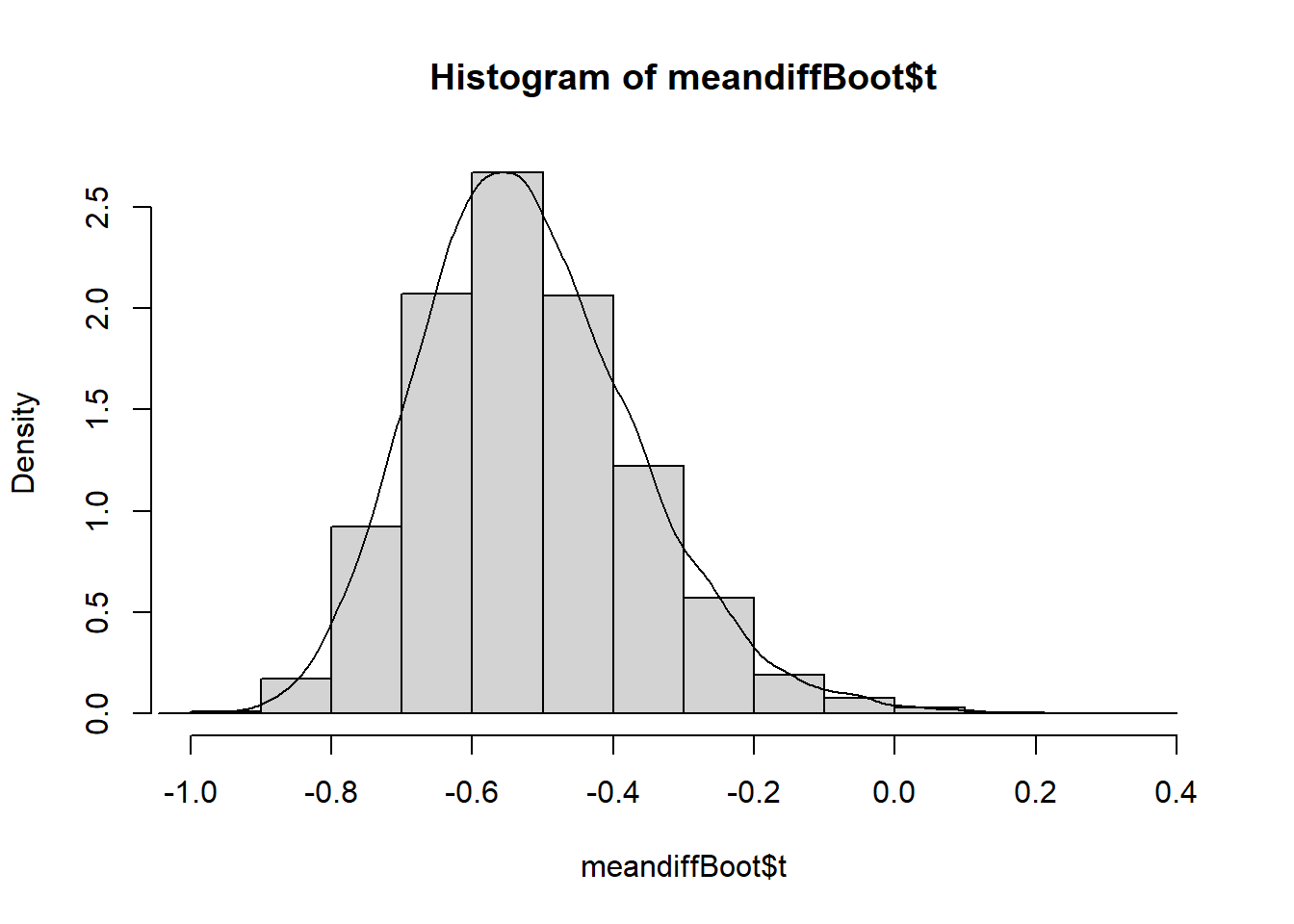
View the summary output and calculate the confidence interval, then add this to your plot above using the abline() function).
Code
meandiffBoot
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot(data = disbud[disbud$Trt == "DB" | disbud$Trt == "LADB",
], statistic = meandiffFun, R = 10000)
Bootstrap Statistics :
original bias std. error
t1* -0.5186062 -0.0004171598 0.1558538Code
lower<--0.5186062-1.96*0.1558538
upper<--0.5186062+1.96*0.1558538
lower[1] -0.8240796Code
upper[1] -0.2131328Code
#plot density (freq=F) histogram of bootstrap results
hist(meandiffBoot$t,freq=F)
#add a smoothed line to show the distribution, remove NAs
lines(density(meandiffBoot$t,na.rm=TRUE))
#add vertical lines with lower and upper confidence limits
abline(v=c(lower,upper))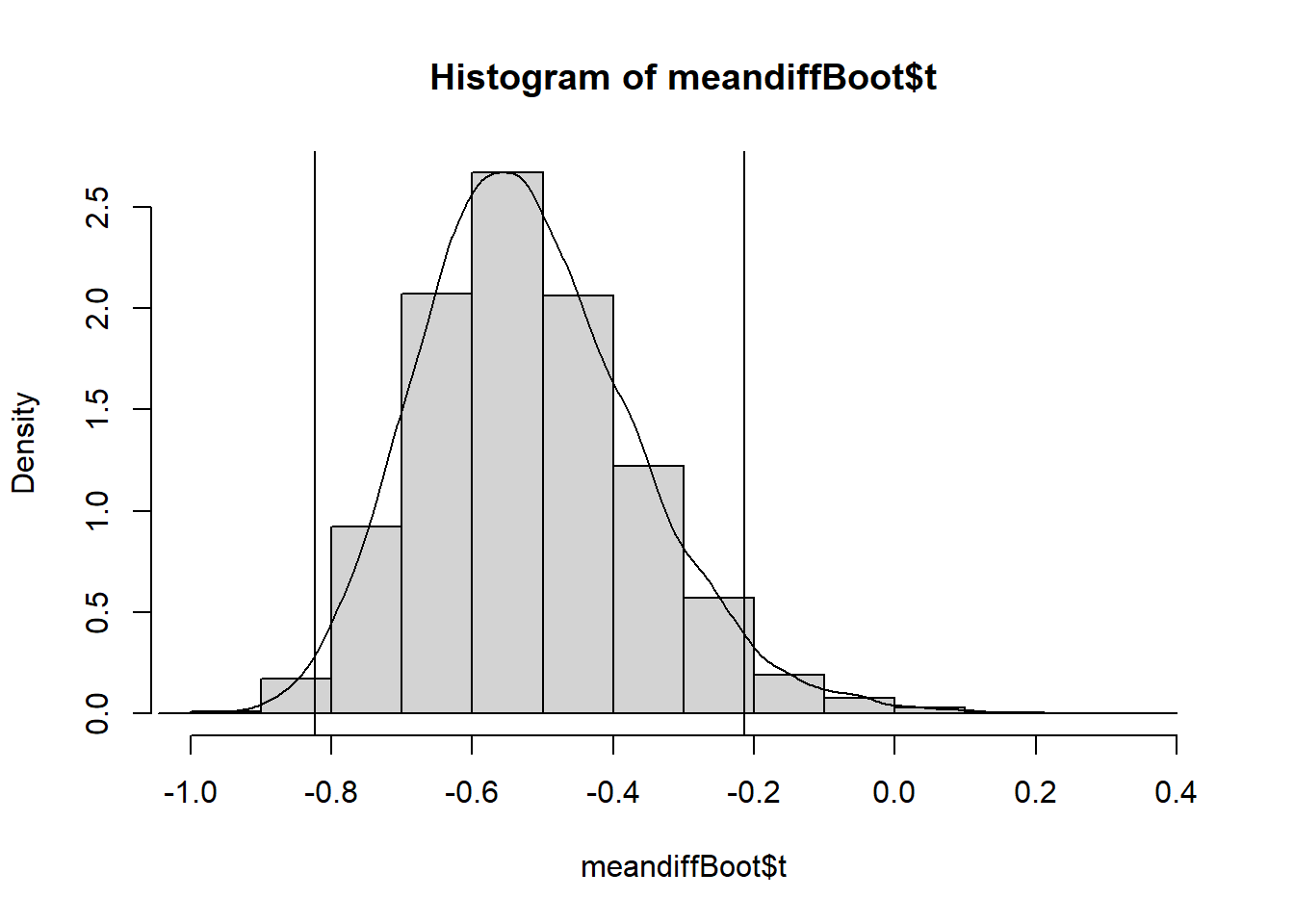
The bootstrapped difference in means follows a near normal distribution around a mean of -0.5186062 with right skew (right side tail). This mean of means matches the difference in means of the disbudding and local anaesthetic with disbudding treatment groups we calculated earlier, and the distribution of bootstrapped difference in mean estimates around it give an indication of the amount of variability in our estimate due to sampling.
The bootstrap confidence interval is narrower than the one calculated earlier and includes only negative values so leads to the same conclusion. There is significant evidence to conclude a difference in mean eye temperature between disbudding and disbudding with local anaesthetic.
As the earlier confidence interval nearly included 0 (upper limit of -0.0818) and bootstrap confidence intervals always differ slightly from those calculated by hand or t.test() this outcome was not necessarily certain.
Construct bootstrap confidence intervals and histograms of the results for the other four differences in means.
7. Simple Linear Regression, ANOVA
An alternative to pairwise bootstrapping for the comparison of means is the Analysis of Variance procedure: ANOVA. This tests for a significant difference between 2 or more groups, and can test for a significant difference between all 4 Trt groups at once.
Fit a linear model for the effect of Trt on ChangeEyeTemp and then carry out ANOVA for this object.
Report your findings. What conclusions can you draw from the results you have obtained so far?
Code
#fitted linear model
disbud.trt<-lm(disbud$ChangeEyeTemp~disbud$Trt)
summary(disbud.trt)
#calculate ANOVA object
anova(disbud.trt)
#residual checks
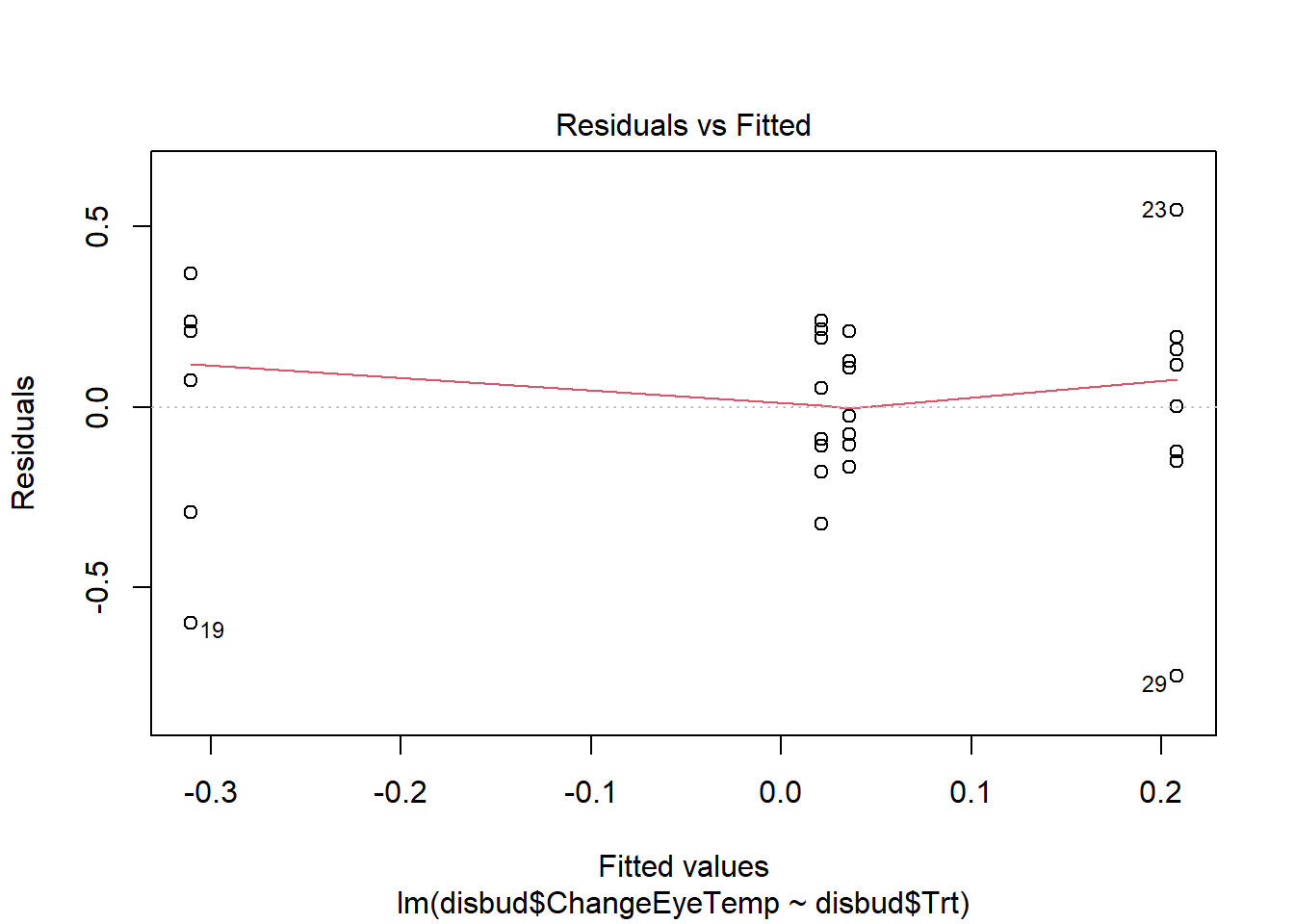
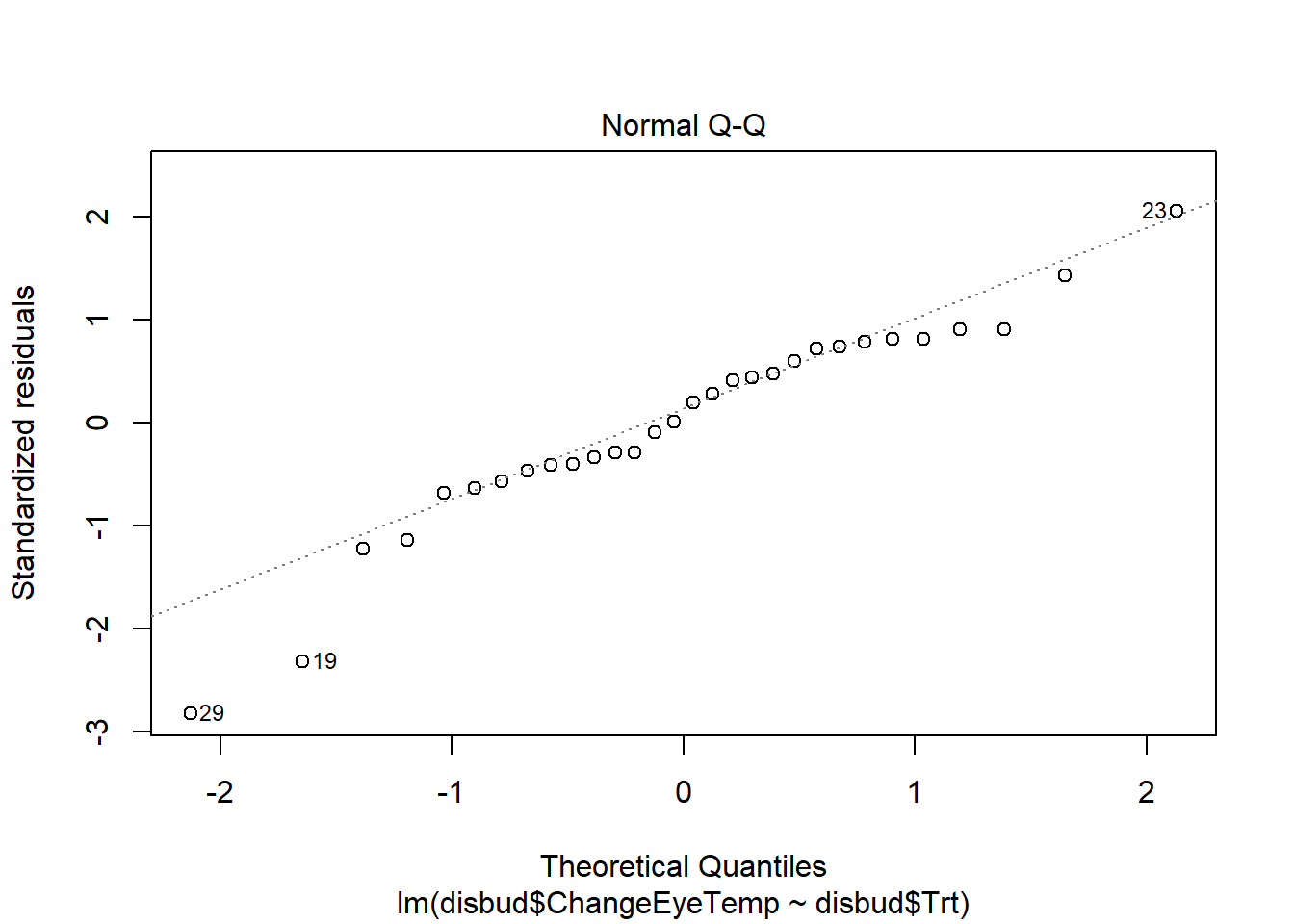
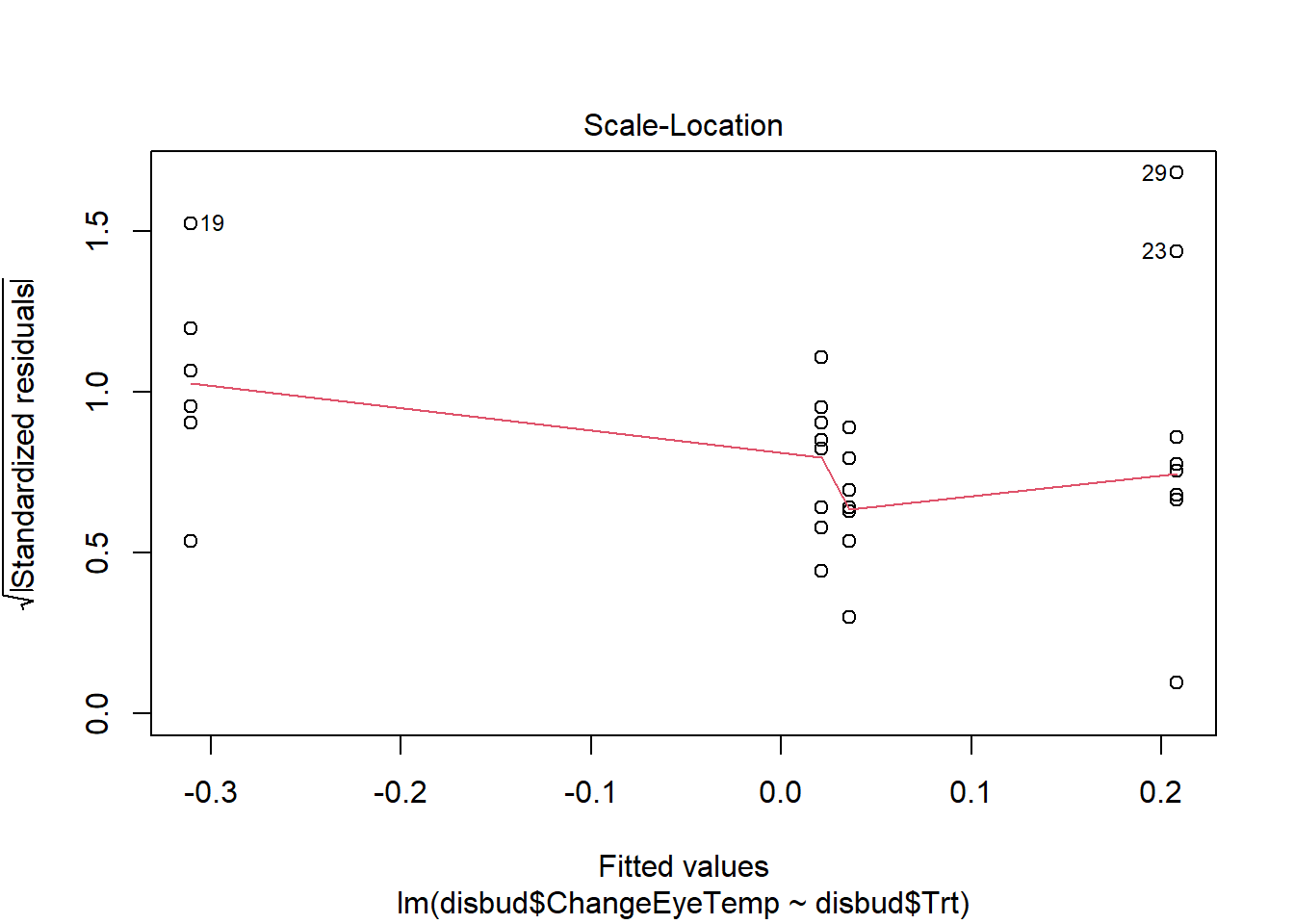
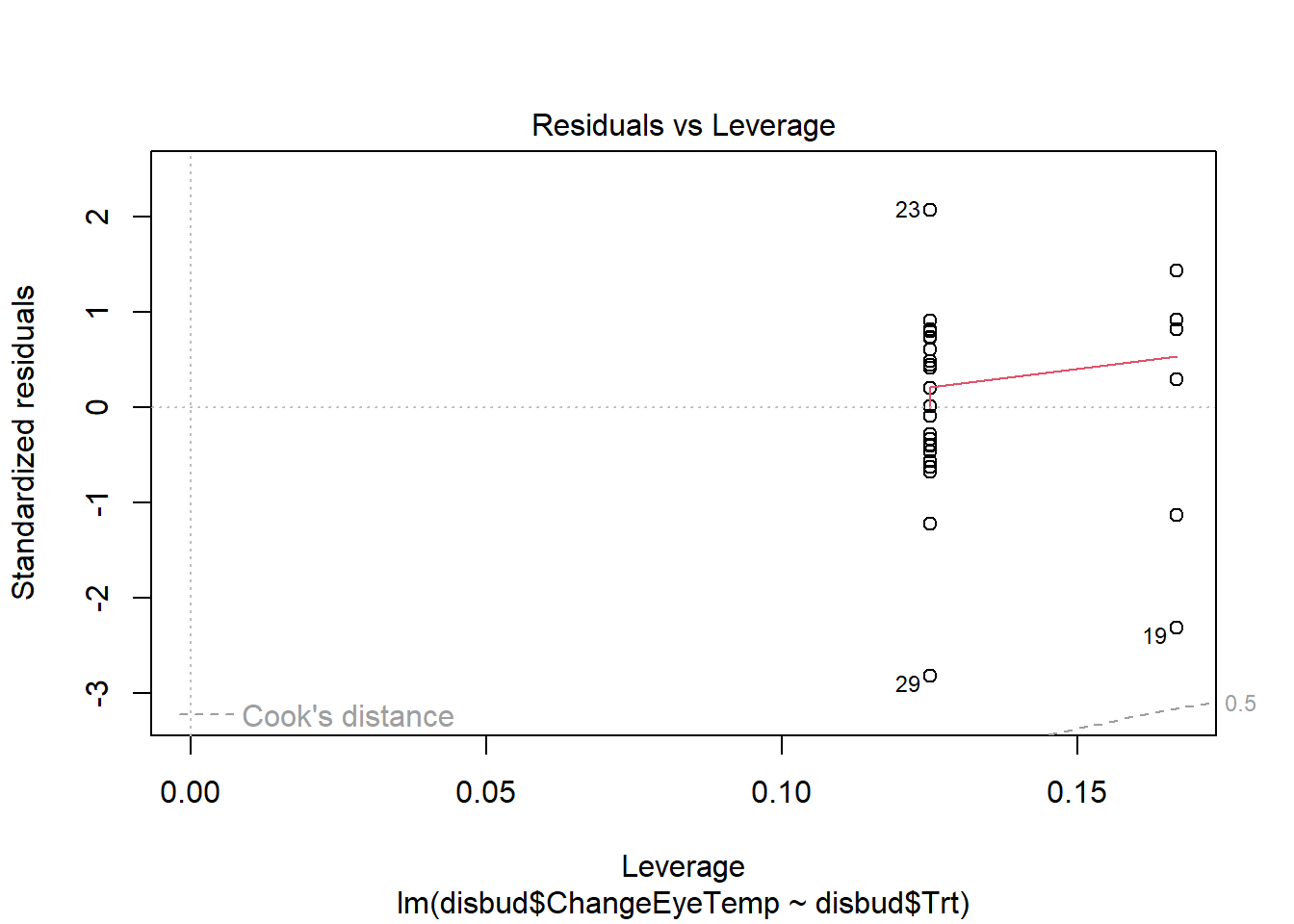
plot(disbud.trt)Code
#fitted linear model
disbud.trt<-lm(disbud$ChangeEyeTemp~disbud$Trt)
summary(disbud.trt)
Call:
lm(formula = disbud$ChangeEyeTemp ~ disbud$Trt)
Residuals:
Min 1Q Median 3Q Max
-0.74582 -0.11896 0.02706 0.19426 0.54555
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.02133 0.09989 0.213 0.8326
disbud$TrtDB -0.33189 0.15259 -2.175 0.0389 *
disbud$TrtLA 0.01441 0.14127 0.102 0.9195
disbud$TrtLADB 0.18671 0.14127 1.322 0.1978
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.2825 on 26 degrees of freedom
Multiple R-squared: 0.3109, Adjusted R-squared: 0.2314
F-statistic: 3.911 on 3 and 26 DF, p-value: 0.01979Code
#calculate ANOVA object
anova(disbud.trt) Analysis of Variance Table
Response: disbud$ChangeEyeTemp
Df Sum Sq Mean Sq F value Pr(>F)
disbud$Trt 3 0.9366 0.312200 3.911 0.01979 *
Residuals 26 2.0755 0.079826
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Code
#residual checks
plot(disbud.trt)